1. Quick Search for the interaction network of individual drug
The users could input a drug generic name or synonym (such as Acetylsalicylic acid, ABC) in the search bar of the home page to query the whole interaction network of the input drug. If the typed terms could match with multiple drug entries in the database, a list of suitable suggestions will be provided to users, with hyperlinks to corresponding full pages. Sometimes the number of matched entries is zero, possibly because of error input or unrecorded drugs. Note that not all the approved drugs are included in the current release, and the interaction networks of some drugs have never been identified. The query can be submitted by clicking the searching icon.
2. Data browser
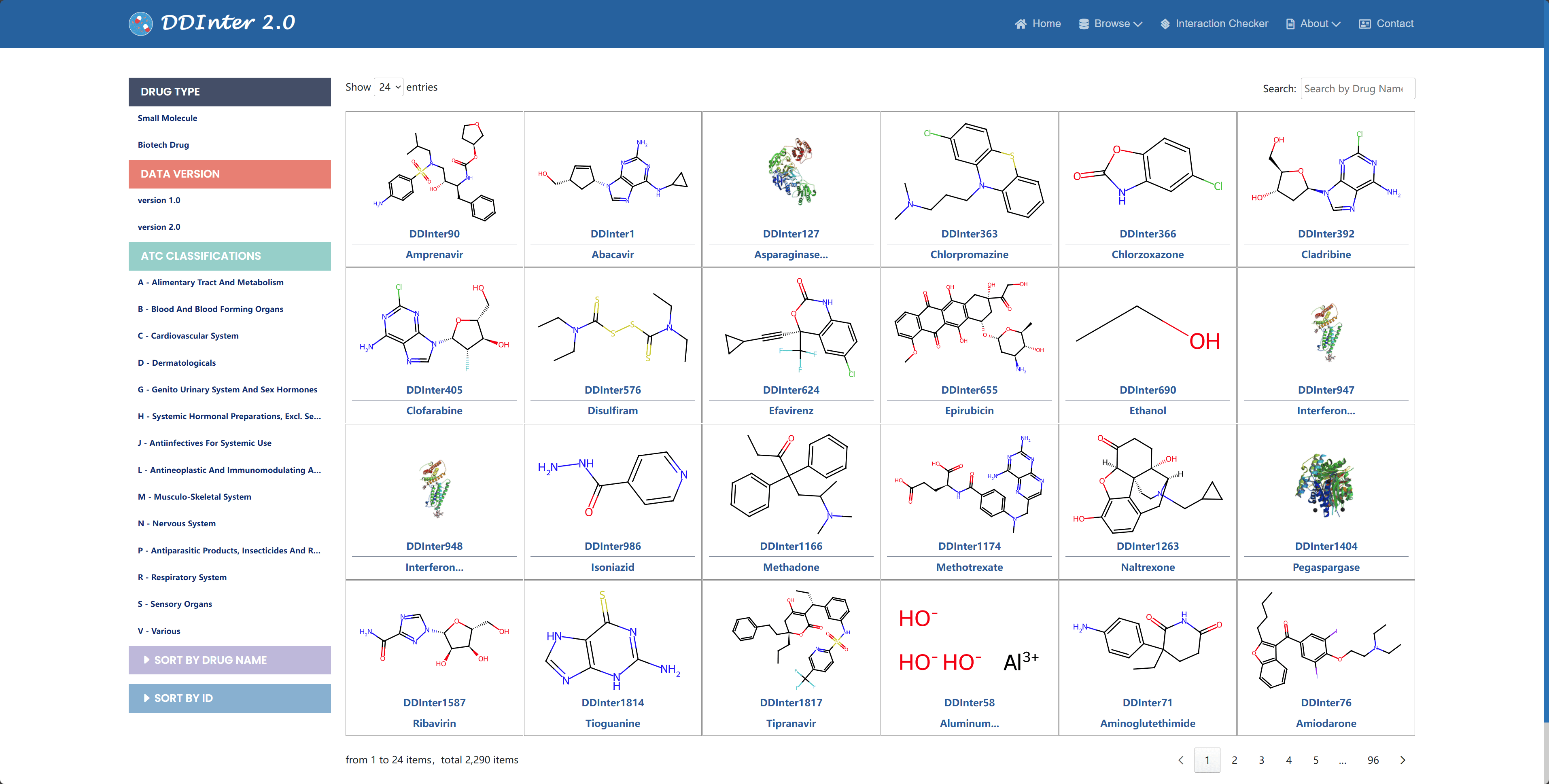
DDInter 2.0 provides comprehensive data browsing capabilities that allow users to query four types of data entries: drug, disease, DDI, and other types of drug interactions (including DFI, DDSI, and therapeutic duplicates), and the system visualizes the chemical information of each drug entity in the form of molecular structure diagrams and assigns unique identifiers to them. In order to facilitate the user to filter and locate drugs of interest, an interactive filtering function is provided on the left side of the page, which supports the user to subset by drug type and therapeutic area, and the system also provides two different sorting methods, namely alphabetical order of drug names and internal numbering order of the database. By clicking on the identifier of a particular drug, users can access its detail page to view basic information, interaction data, and chemical and pharmacological characterization of the drug, as well as external links to other commonly used drug databases to expand on other important information about the drug.
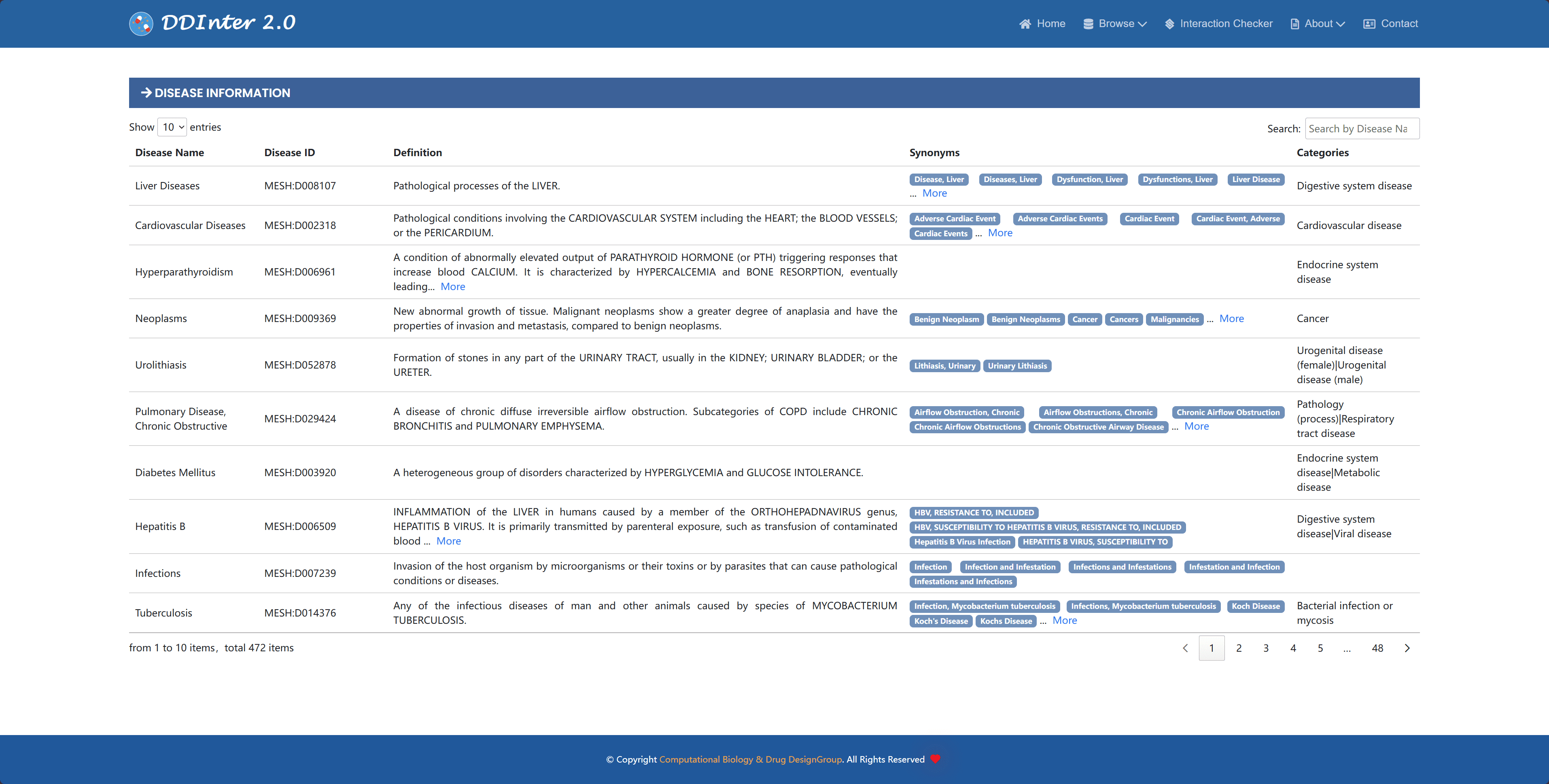
Disease information is presented in a tabular format in the database, including the disease name, MESH number, definition, synonyms, and the disease classification to which it belongs. This structured presentation helps users to quickly understand the key features and hierarchical relationships of the disease.
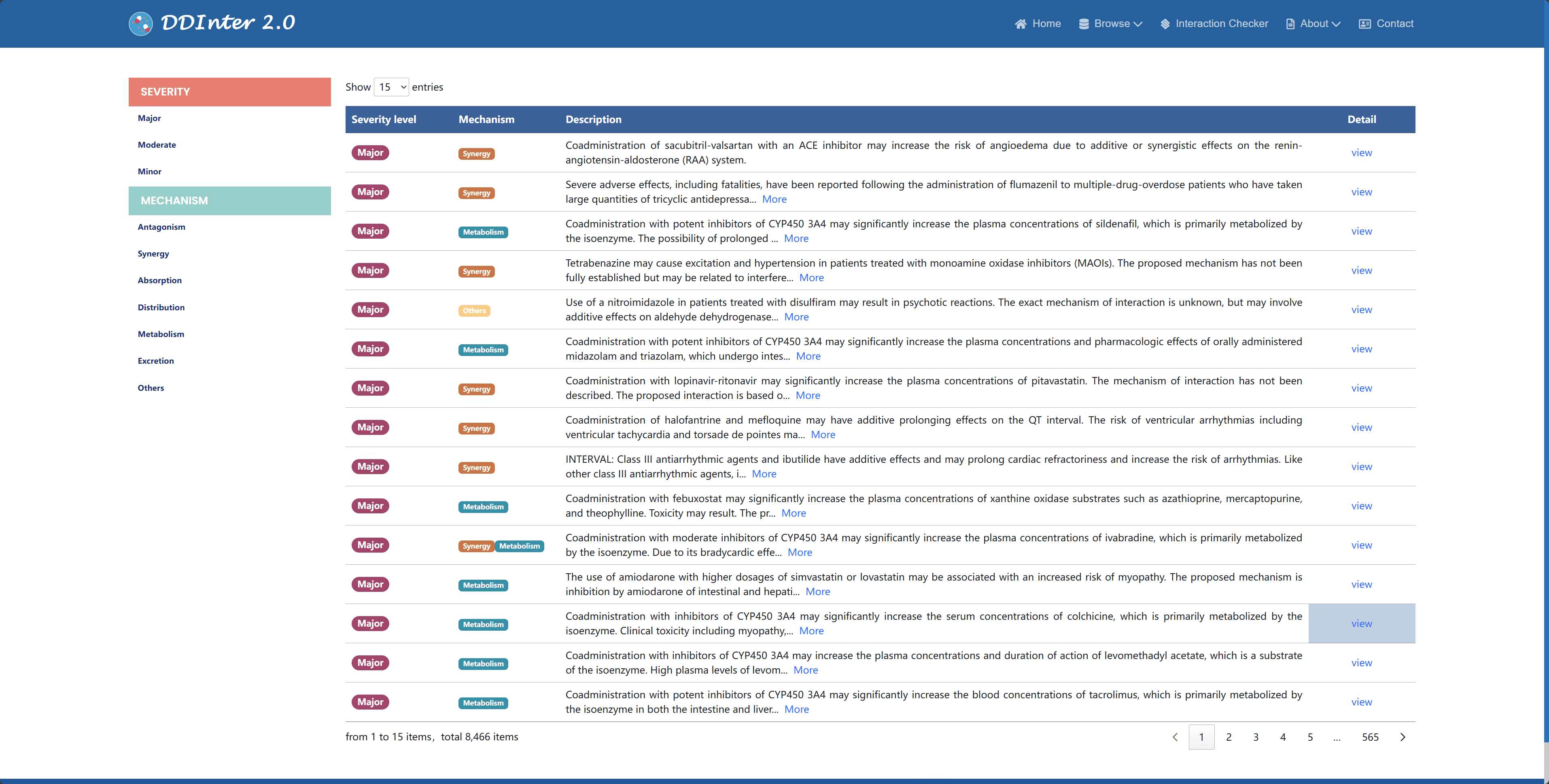
When browsing interaction data, the database provides filters based on severity and mechanism of action to help users narrow down their search and focus on the type of interaction of priority interest. Interaction entities are also presented in a tabular format and can be sorted by drug name or database number. By clicking the "View" button, users can go to the detail page of a specific interaction entry, get a list view of drug-entity pairs, and access specific interactions via hyperlinks. It is important to note that drug entities are sorted by identifier by default, while interaction entities are sorted by risk level to highlight information of higher clinical importance.
3. Drug Details Page Display
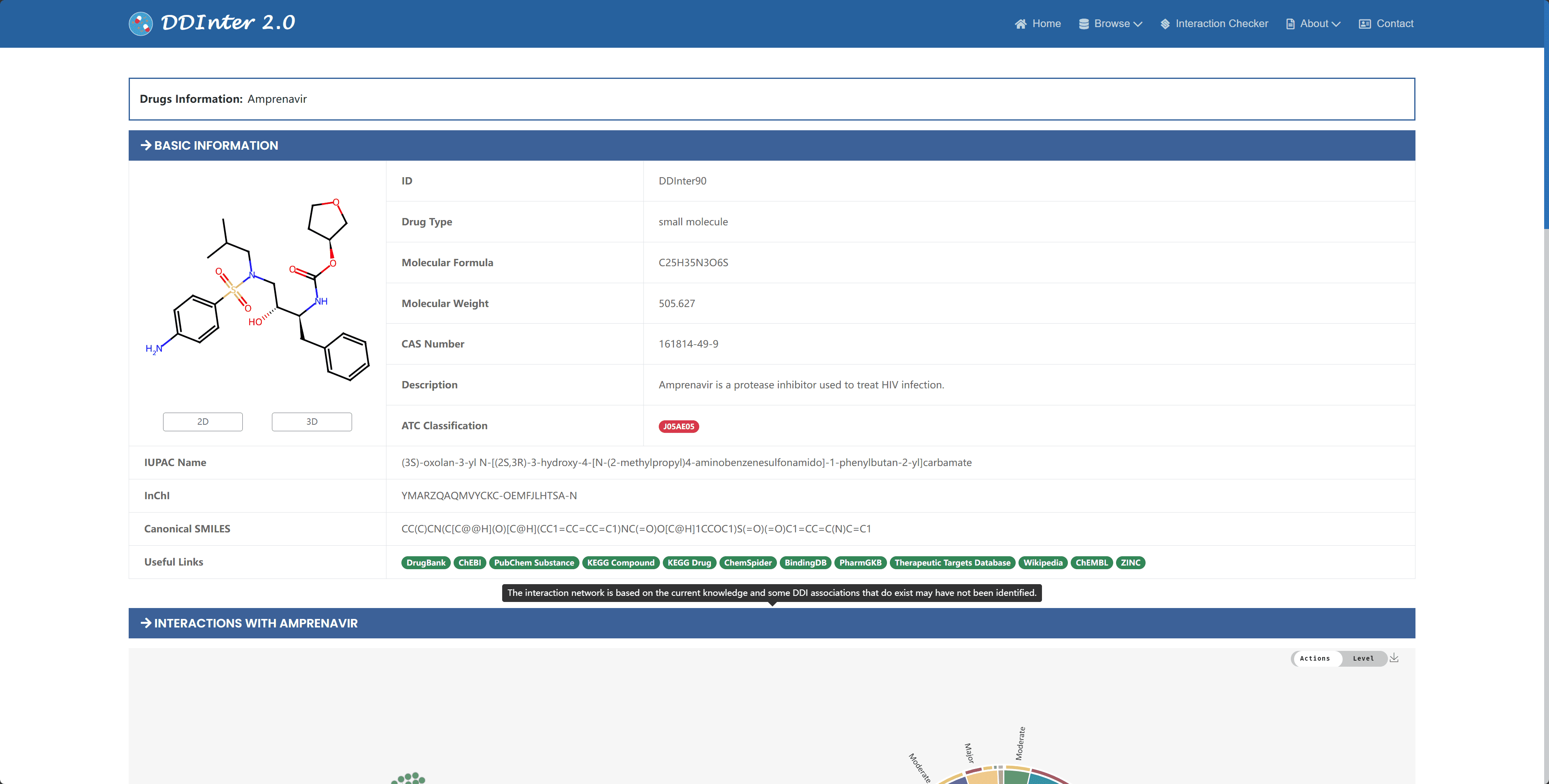
On the drug details page, the information is divided into two main sections: basic information and interaction information. The basic information section includes the chemical and pharmacological characteristics of the drug, such as drug number, type, molecular formula, molecular weight, pharmacological summary, ATC code, external links, and chemical structure representation.
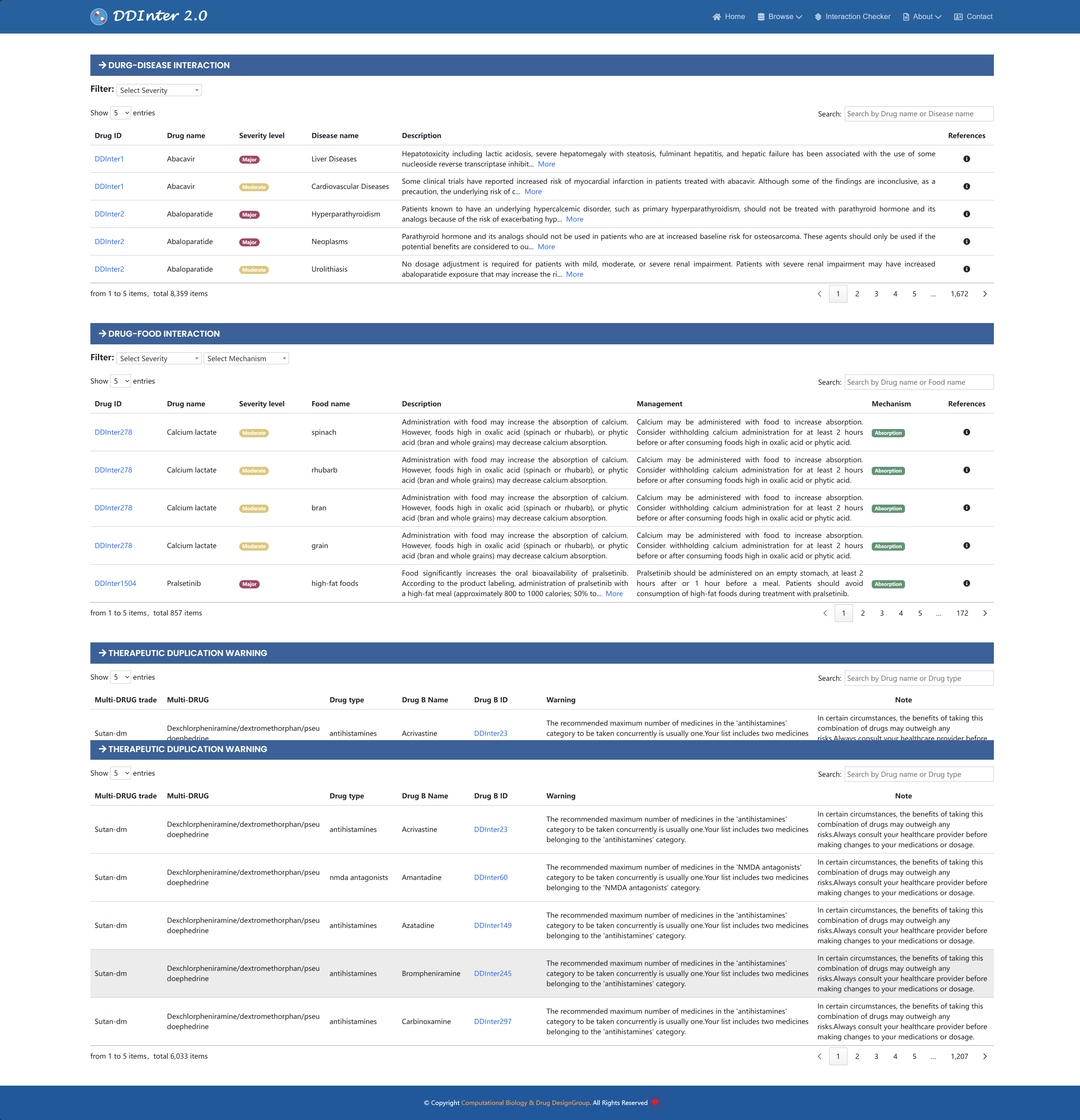
The interaction information section was broken down by interaction type. Among them, DDI information is presented in a tabular format, containing severity, identifier and name of the relevant drug, mechanism of action, and hyperlinks to the detailed DDI page. In the DDI detail page, extended descriptions of interaction mechanisms, such as synergistic toxicity or metabolizing enzyme inhibition, are further provided, as well as corresponding clinical management strategies, such as avoiding co-administration, monitoring for toxicity, adjusting drug dosage, and altering the timing of administration. This detailed annotated information is of great value in guiding clinical decision-making, which can help physicians weigh the risk of DDI and optimize drug treatment regimens. At the same time, the system also provides sources for extracting drug interaction data, making it easy for users to trace the original literature.
Other types of interaction information, such as DDSIs, DFIs, and therapeutic duplicates, are also presented in tabular form following the DDI information. For DDSI, the database provides the risk level, name of the relevant disease, management advice and references; for DFI, the risk level, mechanism of action, name or category of the relevant food, details of the interaction, and management advice; and for therapeutic duplication, the trade name of the compounded preparation, generic name, pharmacologic class, and cautionary statement are given.
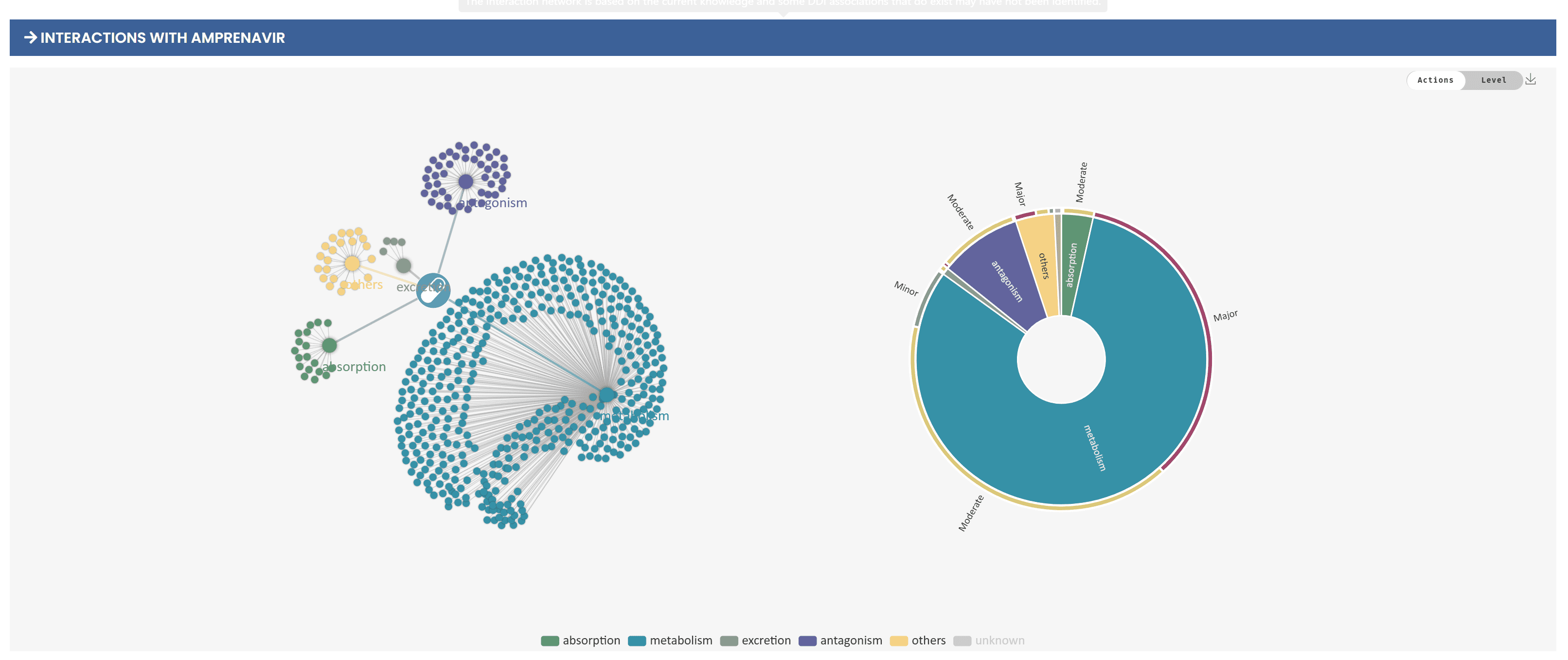
4. Visualization of drug interactions
To help users understand DI information more intuitively, the system provides two visualizations, the relationship graph and the rising sun graph. Users can switch between mechanism of action-based and risk-based views via a switch in the upper right corner of the graph. In the relationship graph, the secondary nodes indicate the mechanism of action or risk level, and there are connections between the interacting drugs and these nodes. Users can view specific interaction descriptions by hovering over the connections and filter out nodes of no interest in clicking on legend entities. The Rising Sun diagrams, on the other hand, depict the distribution of DDIs in a hierarchical manner, and the user can expand further by clicking on each sector to view the next level of information. These graphs support downloading in PNG format, making it easy for users to save and share them. The system also provides filtering and optimization tools to help users narrow down their search and quickly locate the desired interaction information.

5. Drug substitutes and interactions check function
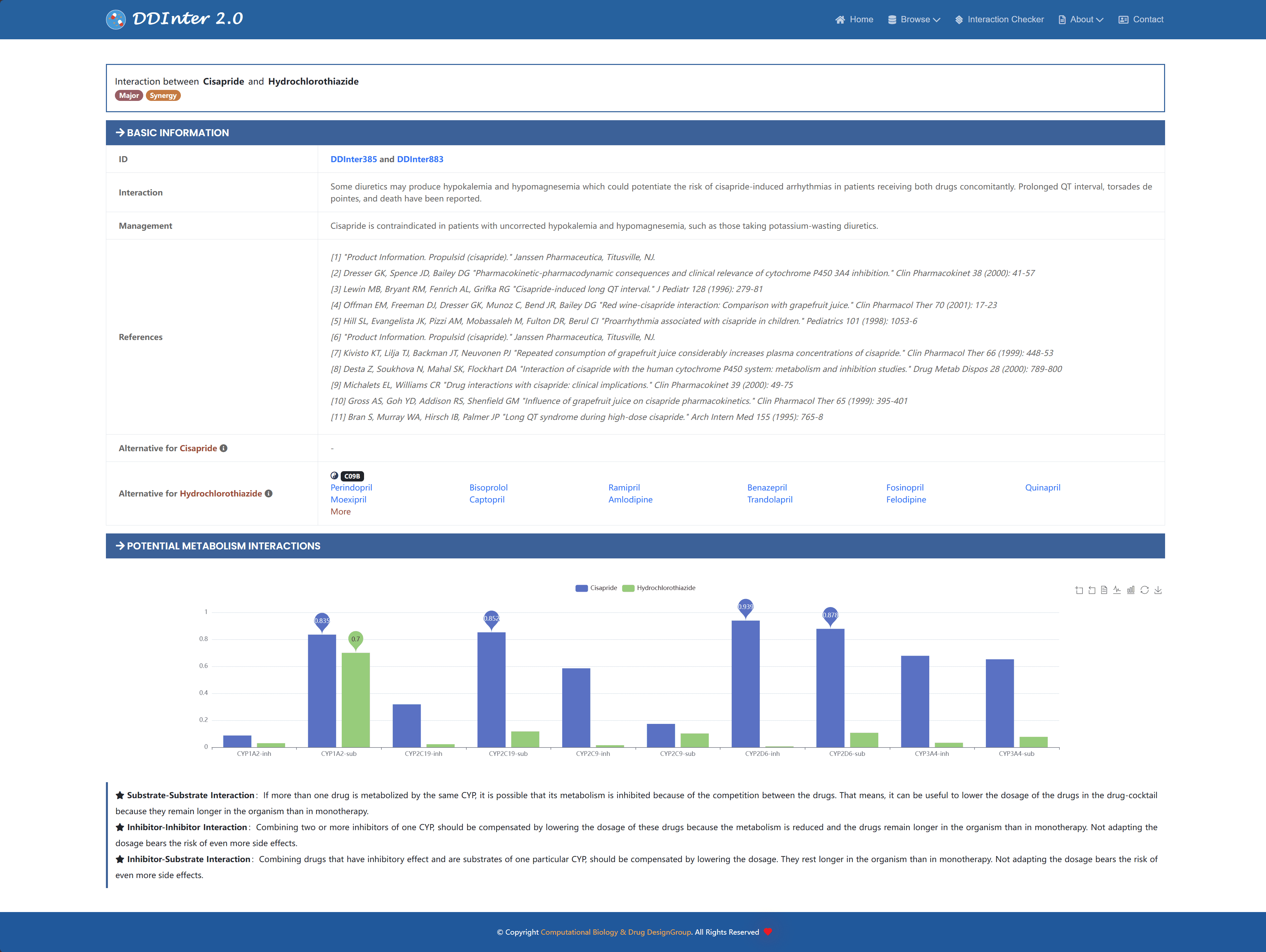
In clinical practice, replacing the original drug with another drug with similar efficacy but lower risk of interaction is a common strategy for managing drug-drug interactions (DDIs). To support this need, the system provides information on potential substitutes for each drug based on the third level (pharmacologic subgroup) of the ATC (Anatomical Therapeutic Chemistry) taxonomy. This feature helps healthcare professionals to quickly find suitable alternatives, reduce the risk of DDIs, and improve patient medication safety.
In addition, the system presents the characteristic predictive values of the drugs in terms of metabolism by the five major CYP isoenzymes in the form of bar charts, which provide a reference for studying potential metabolic interactions that have not yet been detected or summarized. When the predicted value is higher than 0.7, it indicates that the corresponding drug tends to be a substrate or inhibitor of a specific enzyme, thus increasing the probability of a DDI event. Users can browse the specific predicted values, switch the chart type, adjust the zoom ratio, and download the complete visualization results for further analysis and research via the interactive action panel in the top right corner of the page.
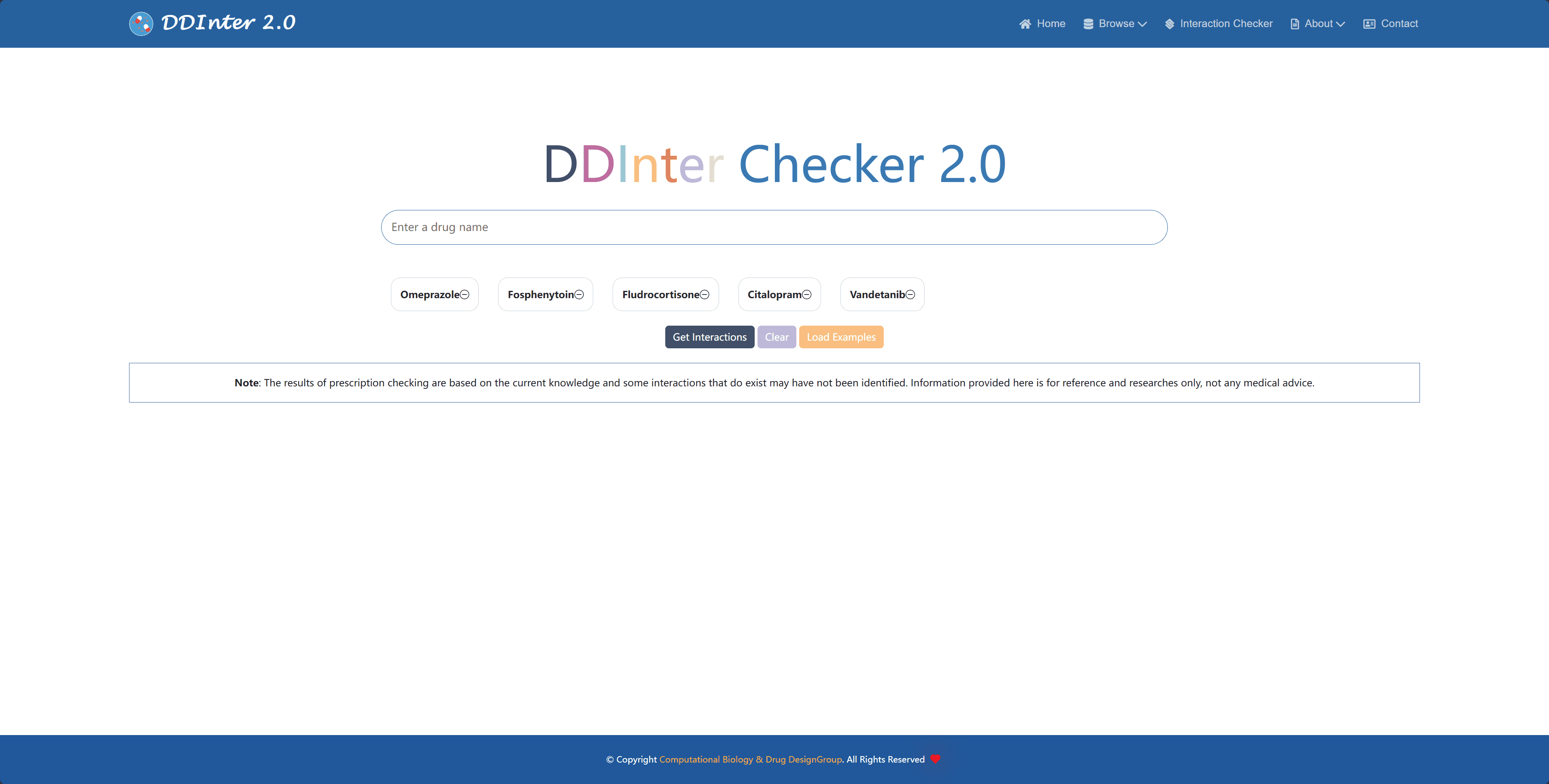
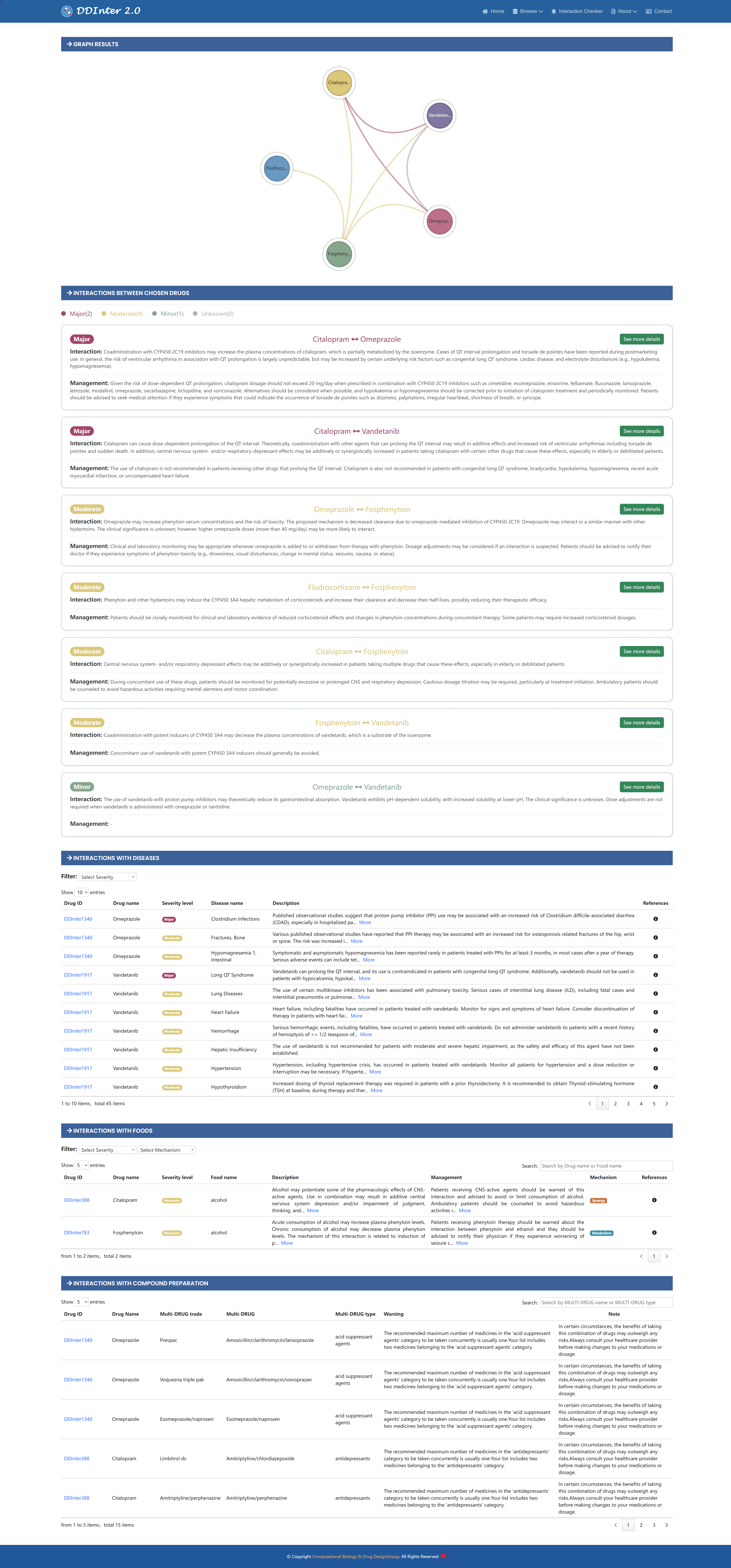
In order to facilitate healthcare professionals to quickly screen prescriptions for potential DDI risks in their daily work, the system provides a dedicated Interaction Checker module that can be accessed directly from the top navigation bar. The module is based on a complete DDI database and supports simultaneous checking of interactions between up to five drugs. The results of the detected DDI associations are presented in the form of a report card, which includes key information such as risk level, interaction mechanism, and management recommendations. At the same time, the system generates a relationship diagram that visualizes the topology of the DDI network between the selected drugs. With simple risk counting statistics and color coding, users can quickly understand the level of danger for each DDI. If more details are needed for a specific DDI entry, the user simply clicks on the "See more details" button and is redirected to the corresponding DDI page for the full summary information.